Gram staining of bacteria is a routine diagnostic method of long standing that can be used for initial diagnoses and to simplify the choice of antibiotics.
It is a simple way to classify bacteria into two classes - Gram-positive and Gram-negative - under a microscope.
In the journal Angewandte Chemie [see below], American researchers have now introduced an improvement to this method: magnetic Gram staining.
This allows for the class-specific, automated, magnetic detection and separation of bacteria.
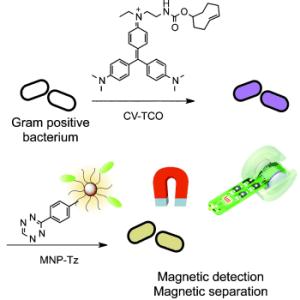
Gram staining was developed about a hundred years ago by Danish bacteriologist Hans Christian Gram. In this technique, bacterial cultures are colored by a stain known as crystal violet, which enters into the murein layer of the bacterial cell walls. Treatment with an iodine-containing solution forms water-insoluble complexes between the crystal violet and iodine. There are two classes of bacteria that differ in the structures of their cell walls. A thick murein layer surrounds one class; the others have only a thin one. Whereas subsequent treatment with ethanol dissolves the stain complex out of the thin murein layer, it remains firmly lodged in the thick murein layers. Bacteria whose stain can be washed away in this manner are classified as Gram-negative; those that remain dark purple are Gram-positive.
Scientists working with Ralph Weissleder at Harvard University in Boston (USA) have now developed Gram staining into a magnetic diagnostic technique. To achieve this, they attached a "molecular hook" to the molecules of crystal violet. With this modified dye, the staining process works just as it does with the original. After staining, however, "eyes" that correspond to the "hooks" are used to attach magnetic nanoparticles to the stain. This makes it easy to quantify the bacteria: nuclear magnetic resonance (NMR) instruments detect the magnetization of the nanoparticles.
It is possible to take an NMR measurement before washing with ethanol to obtain the total number of Gram-positive and Gram-negative bacteria, and again after the washing step to determine the concentration of Gram-positive bacteria.
The advantage of this magnetic detection method is its high sensitivity. It is possible that samples could be directly magnetized and measured without prior purification or culture of the bacteria. By using the simple but sensitive miniaturized micro-NMR instruments developed by this research group, fast and sensitive on-the-spot diagnosis is conceivable. In addition, the magnetization could be used for the separation of bacteria from the sample.
About the Author
Dr. Weissleder is a Professor at Harvard Medical School, Director of the Center for Systems Biology at Massachusetts General Hospital (MGH), and Attending Clinician at MGH. His research interests include the development of novel molecular imaging techniques, tools for detection of early disease detection, development of nanomaterials for sensing and systems analysis. He is a member of the US National Academies Institute of Medicine.
Further Information:
Dr. Ghyslain Budin, Dr. Hyun Jung Chung, Prof. Hakho Lee, Prof. Ralph Weissleder:
A Magnetic Gram Stain for Bacterial Detection.
In: Angewandte Chemie International Edition; published online 28 June 2012, DOI 10.1002/anie.201202982
Source: Angewandte Chemie International Edition, press release 27/2012
Last update: 15.07.2012
Perma link: https://www.internetchemistry.com/news/2012/jul12/magnetic-gram-staining.php
More chemistry: index | chemicals | lab equipment | job vacancies | sitemap
Internetchemistry: home | about | contact | imprint | privacy
© 1996 - 2023 Internetchemistry
